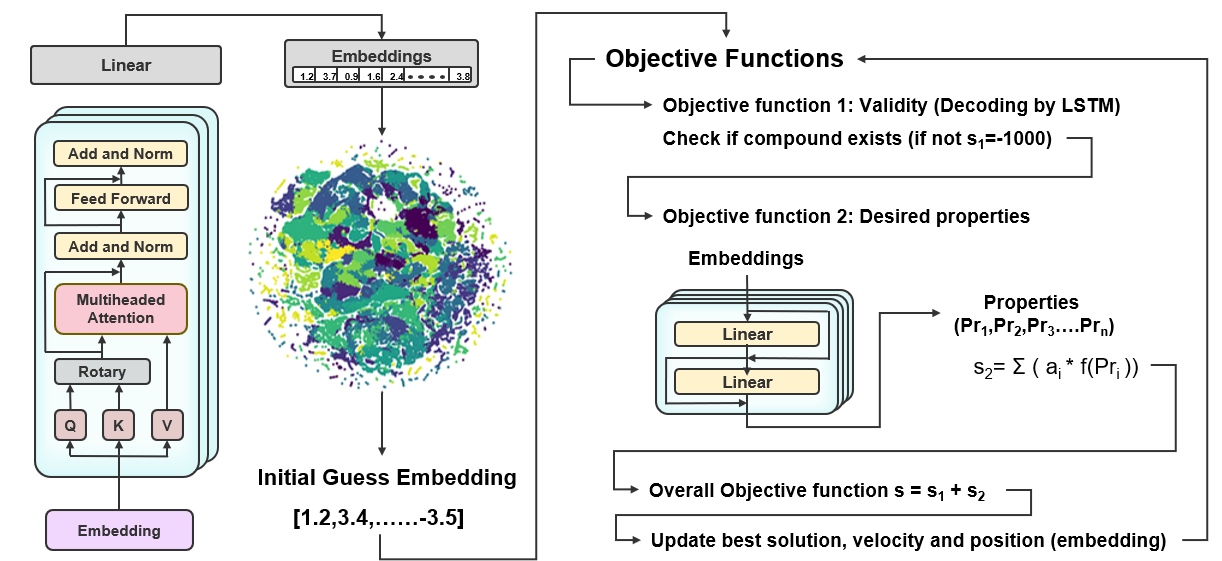
Optimizing drug-like molecules traditionally relies on trial-and-error approaches or brute-force computational searches, making it difficult to explore chemical space efficiently. This project introduces a latent space optimization framework that utilizes transformer-generated molecular embeddings to refine molecular properties and identify optimal drug candidates.
Highlights of this approach:
- Optimization in structured latent space – Instead of optimizing raw molecular descriptors, the model refines properties within a learned chemical space.
- Efficient exploration of molecular candidates – Identifies compounds with desirable pharmacokinetic profiles, balancing bioavailability, metabolic stability, and toxicity.
- Seamless integration with predictive models – Connects molecular design and property prediction, reducing the need for exhaustive screening.
This data-driven approach streamlines molecular optimization, reducing reliance on computationally expensive searches and enabling the more efficient discovery of novel drug candidates.