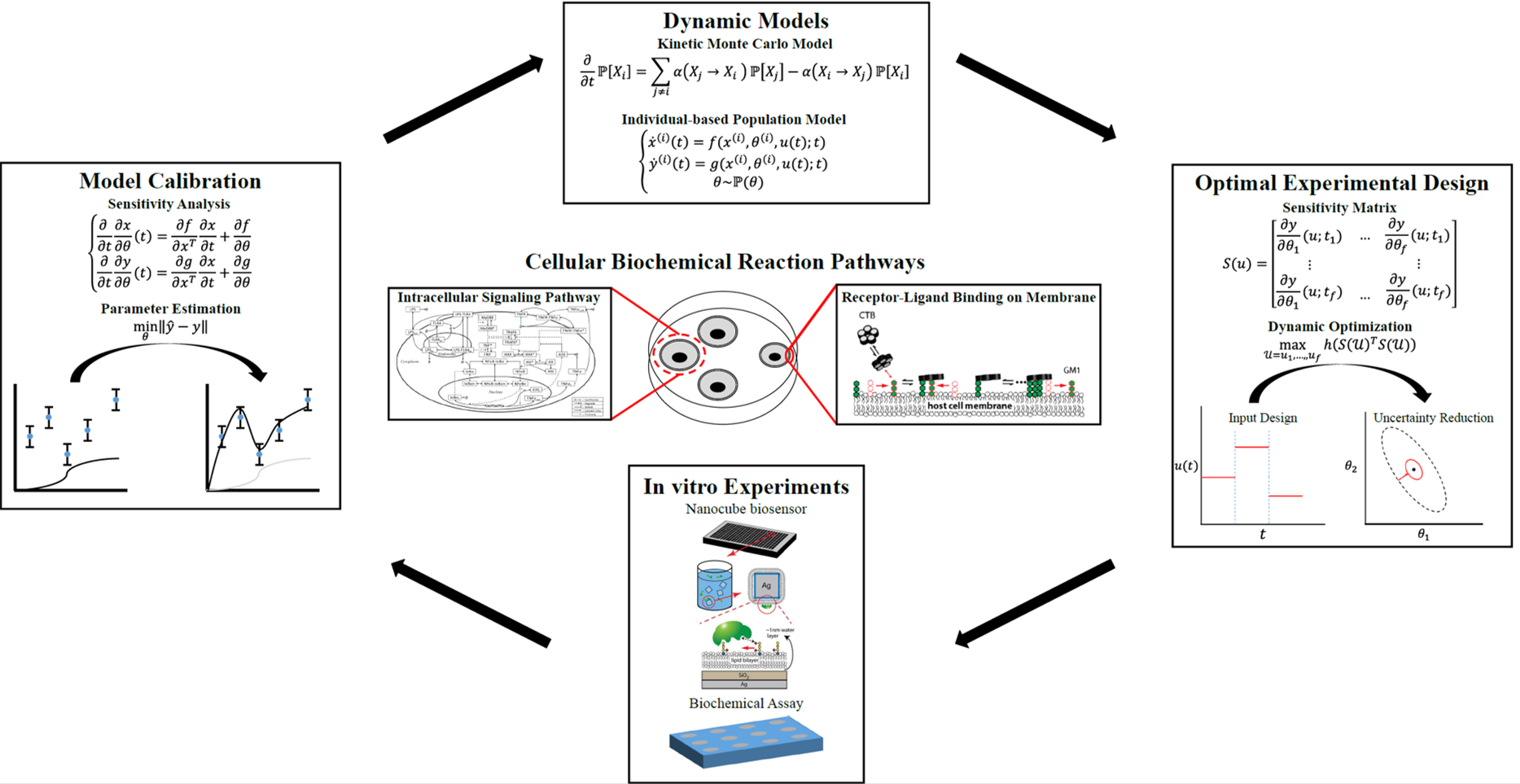
 When a cell perceives an external stimulus, multiple biochemical reaction pathways are triggered simultaneously in order for the cell to respond to the stimulus by changing the activities and expression levels of intracellular molecules. Therefore, the quantitative characterization of the biochemical reaction pathways is critical in understanding the dynamic behaviors of the cells. With the rapid improvements in experiment techniques, it has become easier for researchers to measure the dynamics of a reaction pathway. However, how to process and interpret these data still remains a challenge for researchers. To this end, mathematical modeling has become attractive since it can be used to integrate datasets from diverse sources to verify existing hypotheses. At the same time, the model-based optimal design of experiment techniques can be used to guide future experiments to test alternative hypotheses so that the information from future experiments will be maximized. Currently, our group is developing systematic approaches to integrate diverse and complex datasets through mathematical modeling, sensitivity analysis, and parameter estimation so that the resulting model can be served as a surrogate of the real cellular process to test hypotheses and design an optimal experiment to validate new ones.
When a cell perceives an external stimulus, multiple biochemical reaction pathways are triggered simultaneously in order for the cell to respond to the stimulus by changing the activities and expression levels of intracellular molecules. Therefore, the quantitative characterization of the biochemical reaction pathways is critical in understanding the dynamic behaviors of the cells. With the rapid improvements in experiment techniques, it has become easier for researchers to measure the dynamics of a reaction pathway. However, how to process and interpret these data still remains a challenge for researchers. To this end, mathematical modeling has become attractive since it can be used to integrate datasets from diverse sources to verify existing hypotheses. At the same time, the model-based optimal design of experiment techniques can be used to guide future experiments to test alternative hypotheses so that the information from future experiments will be maximized. Currently, our group is developing systematic approaches to integrate diverse and complex datasets through mathematical modeling, sensitivity analysis, and parameter estimation so that the resulting model can be served as a surrogate of the real cellular process to test hypotheses and design an optimal experiment to validate new ones.
Specifically, we are developing 1) a kinetic Monte Carlo simulation framework to simulate the dynamics of glycan-lectin binding processes on a cellular member to study the host-pathogen interactions, 2) an individual-based population modeling framework to model and explore the origins of cell-to-cell variabilities in the single-cell measurements, and 3) a new optimal experimental design algorithm to guide the future experiments considering the practical constraints and the potential model mismatch.
Literature:
D. Lee, Y. Ding, A. Jayaraman, and J. S. Kwon, “Mathematical modeling and parameter estimation of intracellular signaling pathway: application to LPS-induced NFκB activation and TNFα production in macrophages,” Processes, 2018, 6(3), 21 (Feature Paper)
D. Lee, A. Mohr, J. S. Kwon, and H. Wu, “Kinetic Monte Carlo modeling of multivalent binding of CTB proteins with GM1 receptors,” Comp. & Chem. Eng.,2018, 118, 283-295.
H. Choi, D. Lee, A. Singla, J. S. Kwon, and H. Wu, “The influence of heteromultivalency on lectin–glycan binding behavior,” Glycobiology 2019, 29 (5), 397-408 (Cover page of the “Official Journal of the Society for Glycobiology”).